Publications
Lab members in bold
Preprints
Brand CM; White FJ; Ting N; Webster TH. 2020. Soft sweeps predominate recent positive selection in bonobos (Pan paniscus) and chimpanzees (Pan troglodytes). bioRxiv 2020.12.14.422788.
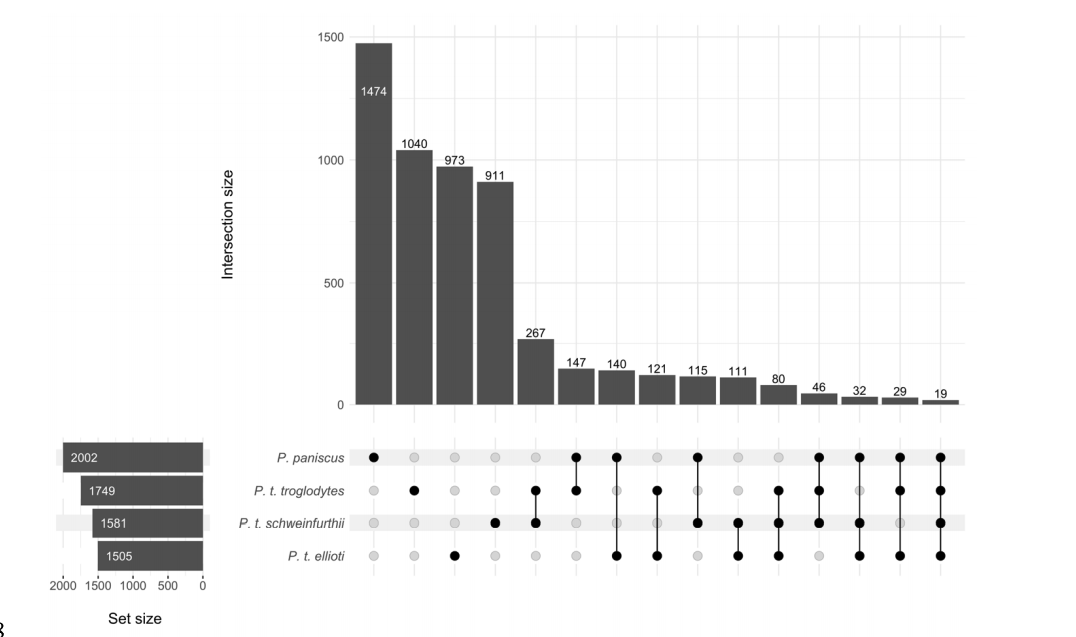
-
Access the paper
- Article Link
- Biorxiv Preprint: 422788
Webster TH; Guevara EE; Lawler RR; Bradley BJ. 2018. Successful exome capture and sequencing in lemurs using human baits. bioRxiv 490839.
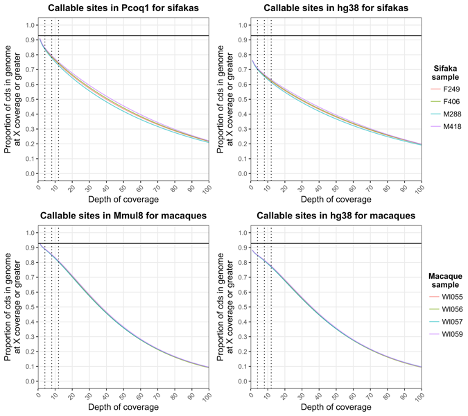
-
Access the paper
- Article Link
- Biorxiv Preprint: 490839
- Code: https://github.com/thw17/Sifaka_assembly
- Online Datasets:
Webster TH*; Golby GA*; Wilson Sayres MA; Kusumi K. 2018. Improved draft of the Mojave Desert tortoise genome, Gopherus agassizii, version 1.1. PeerJ Preprints 3266.

-
Access the paper
- Article Link
- PeerJ Preprint: 3266
- Code: https://github.com/thw17/G_agassizii_reference_update
- Online Datasets: Additional Link:
- NCBI Genome Assembly
2024
Brand CM; Kuang S; Gilbertson EN; McArthur E; Pollard KS; Webster TH; Capra JA. 2024. Sequenced-based machine learning reveals 3D genome differences between bonobos and chimpanzees. Genome Biology and Evolution 16: evae210.
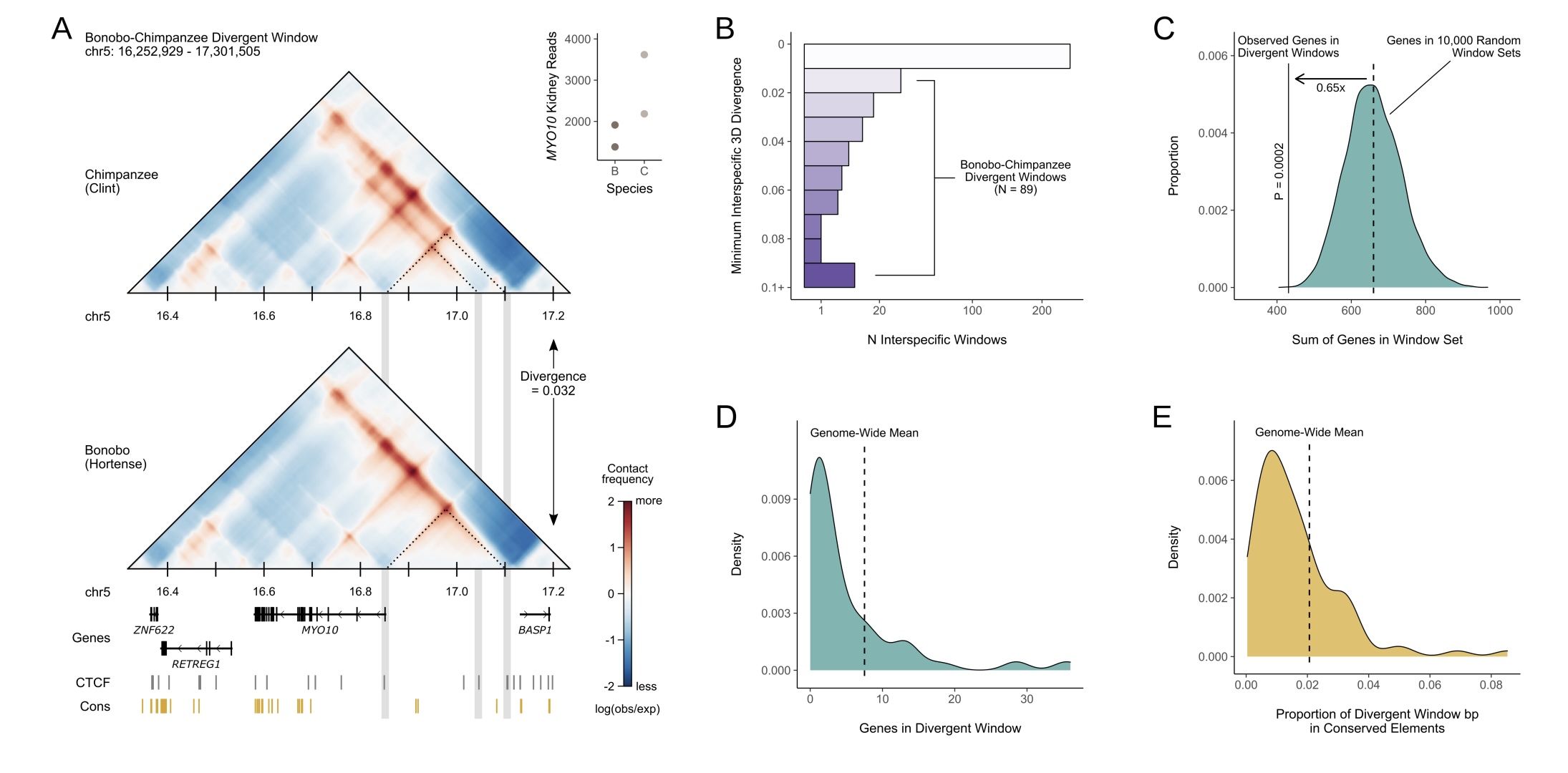
-
Access the paper
- Article Link
- Biorxiv Preprint: 564272
Webster TH; Vannan A; Pinto BJ; Denbrock G; Morales M; Dolby GA; Fiddes IT; DeNardo DF; Wilson MA. 2024. Lack of dosage balance and incomplete dosage compensation in the ZZ/ZW Gila monster (Heloderma suspectum) revealed by de novo genome assembly. Genome Biology and Evolution 16: evae018.
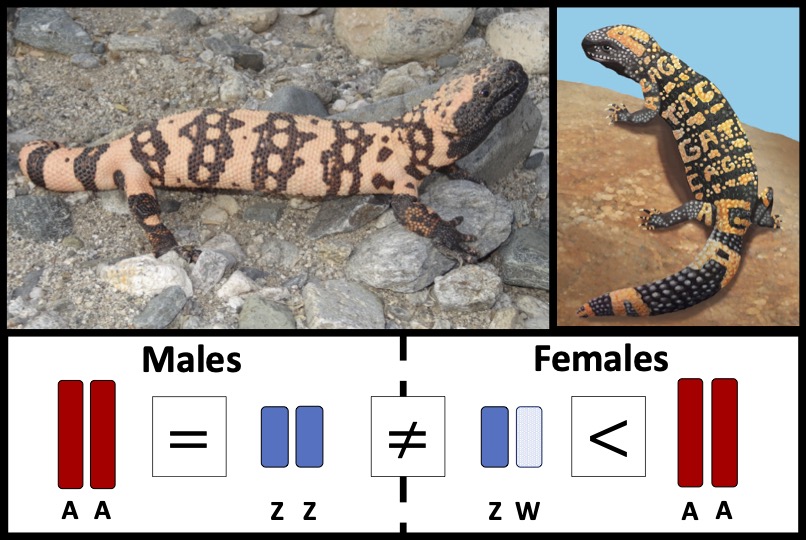
-
Access the paper
- Article Link
- Biorxiv Preprint: 538436
2023
Cotter DJ; Webster TH; Wilson MA. 2021. Genomic and demographic processes differentially influence genetic variation across the X chromosome. bioRxiv 2021.01.31.429027
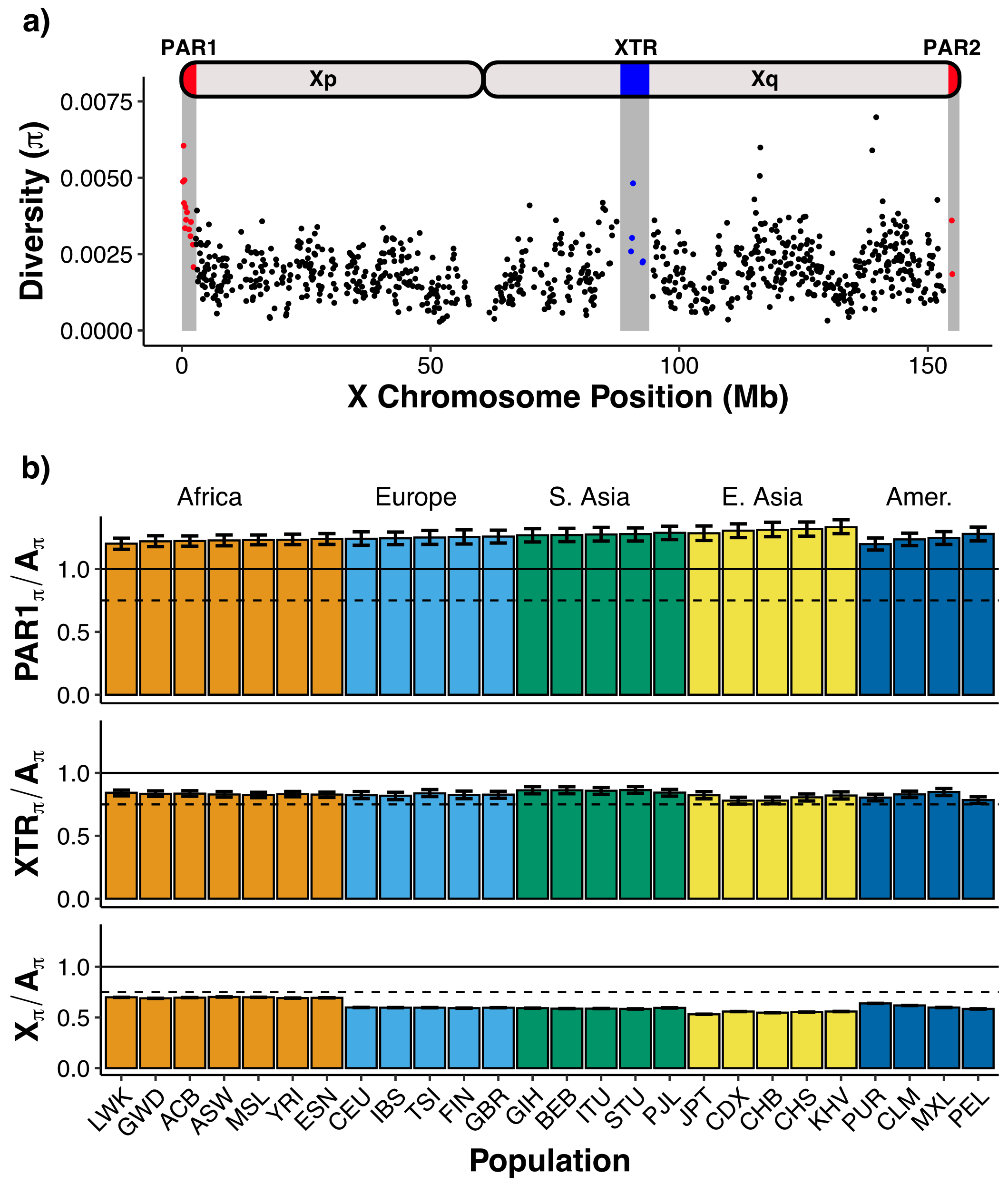
-
Access the paper
- Article Link
- Biorxiv Preprint: 429027
2022
Kollath DR; Dolby GA; Webster TH; Barker BM. 2023. Characterizing fungal communities on Mojave desert tortoises (Gopherus agassizii) in Arizona to uncover potential pathogens. Journal of Arid Environments 219: 105089.
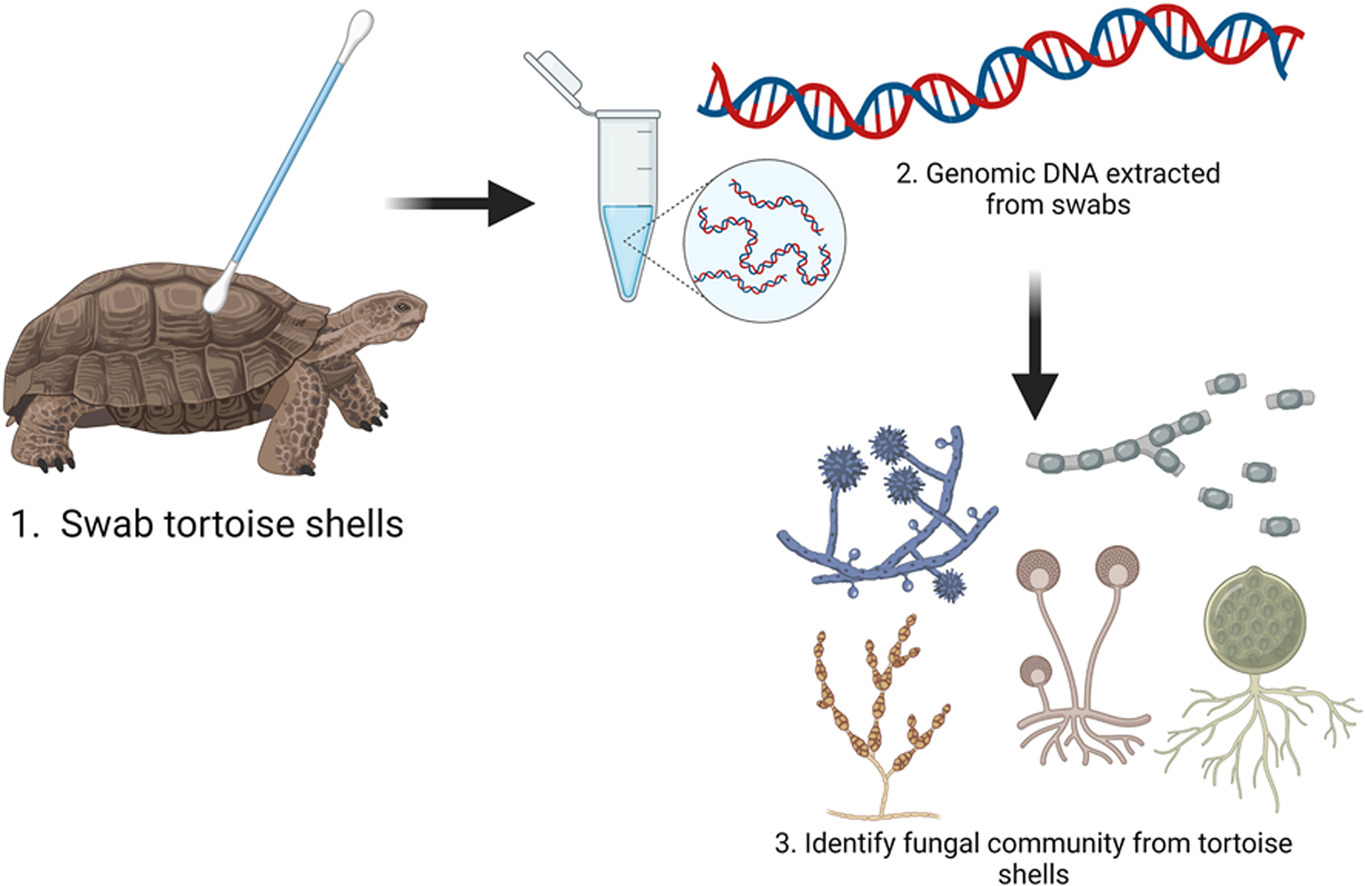
-
Access the paper
- Article Link
Byrne H; Webster TH; Brosnan SF; Izar P; Lynch JW 2021. Signatures of adaptive evolution in platyrrhine primate genomes. Proceedings of the National Academy of Sciences 119: e2116681119.
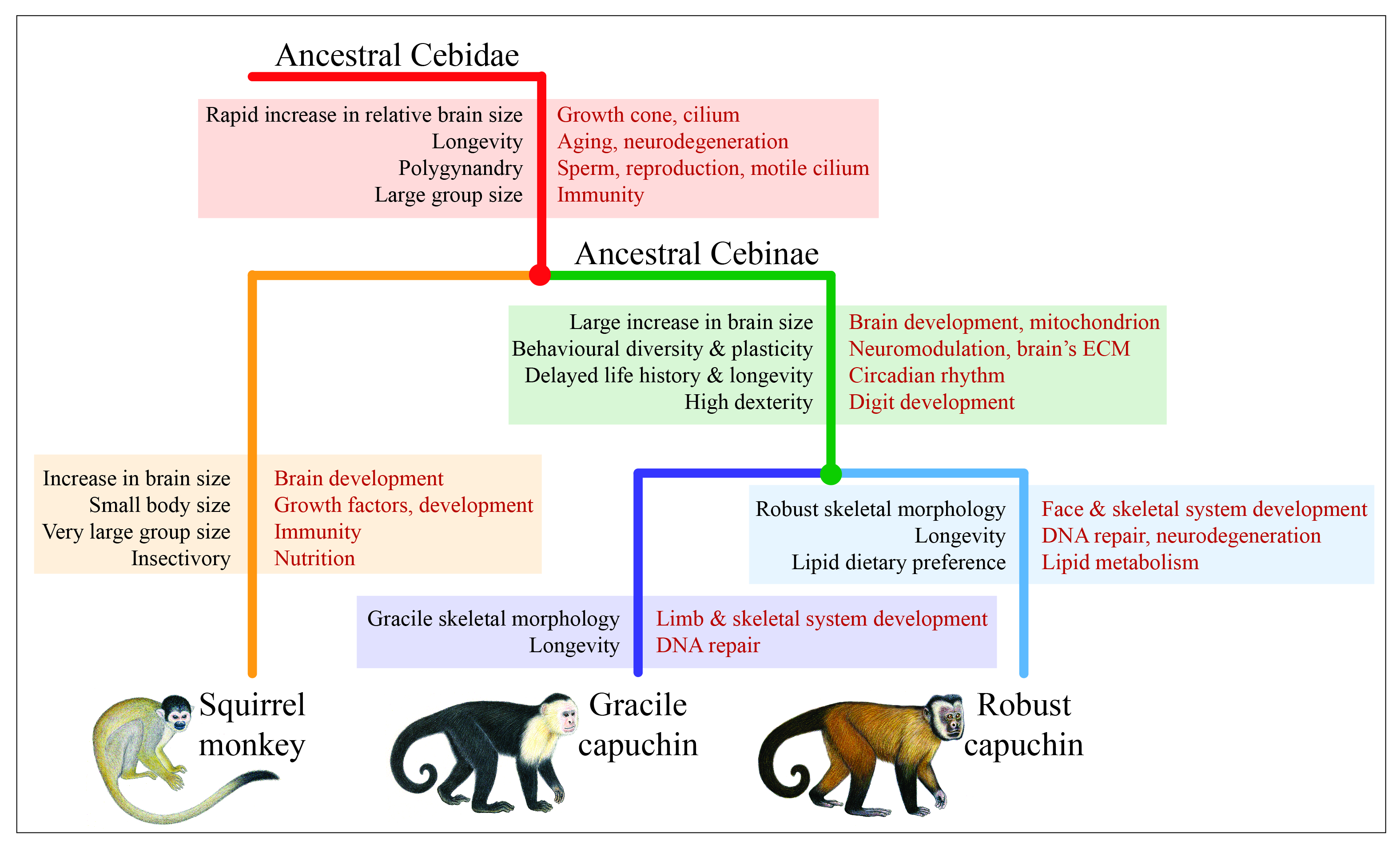
-
Access the paper
- Article Link
- Biorxiv Preprint: 456368
Brand CM; White FJ; AR Rogers; Webster TH. 2022. Estimating bonobo (Pan paniscus) and chimpanzee (Pan troglodytes) evolutionary history from nucleotide site patterns. Proceedings of the National Academy of Sciences 119: e2200858119.
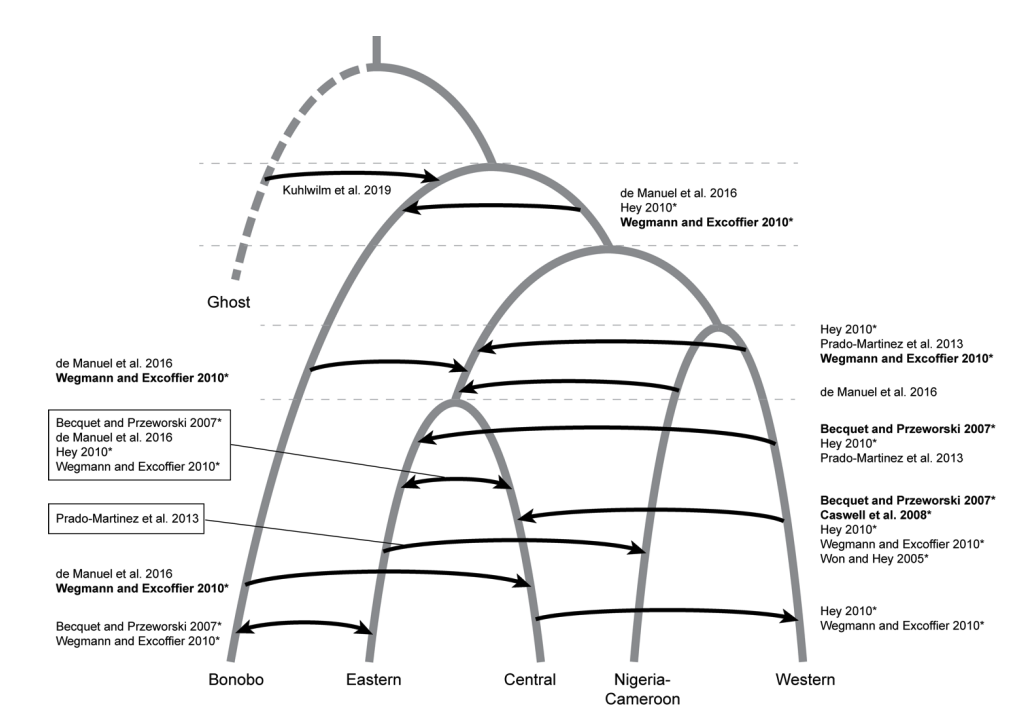
-
Access the paper
- Article Link
- Biorxiv Preprint: 475438
2021
22) Lasisi T; Zaidi AA; Webster TH; Stephens NB; Routch K; Jablonski NG; Shriver MD. 2020. High-throughput phenotyping methods for quantifying hair fiber morphology. Scientific Reports 11: 11535.
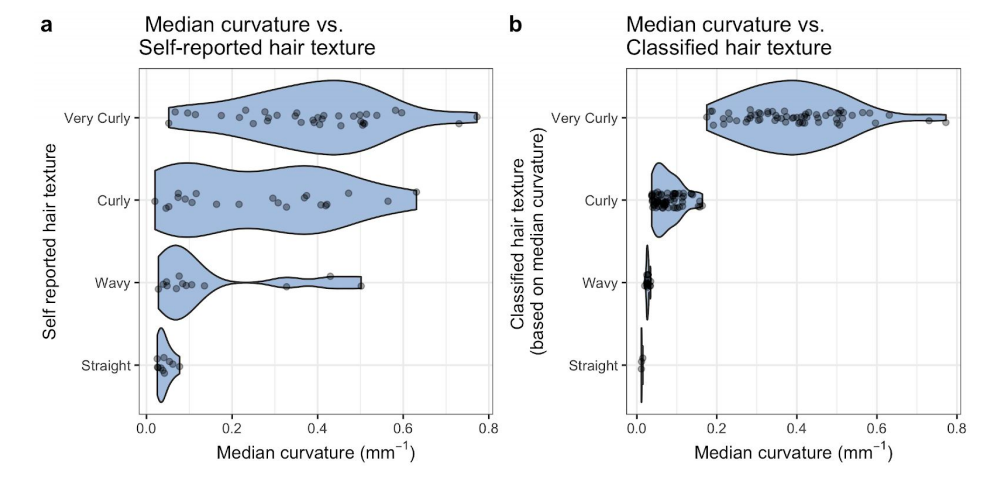
-
Access the paper
- Article Link
- Biorxiv Preprint: 392191 Additional Link:
- Fibermorph software
21) Phung TN; Webster TH; Lenkiewicz E; Malasi S; Adreozzi M; McCullough AE; Anderson KS; Pockaj BA; Wilson MA; Barrett MT. 2021. Unique evolutionary trajectories of breast cancers with distinct genomic and spatial heterogeneity. Scientific Reports 11: 10571.
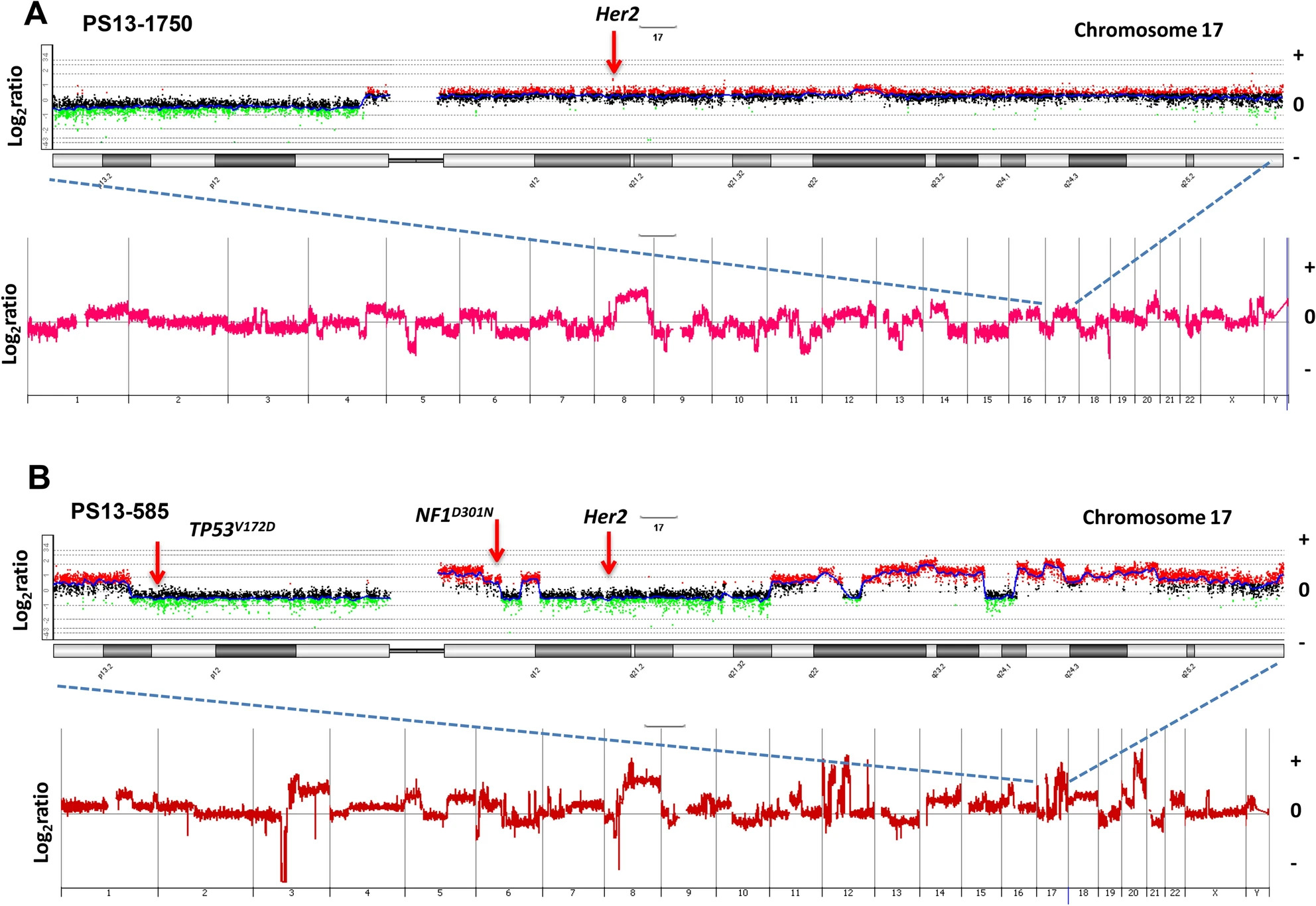
-
Access the paper
- Article Link
20) Guevara EE; Webster TH; Lawler RR; Bradley BJ; Green LK; Ranaivonasy J; Ratsirarson J; Harris RA; Liu Y; Murali S; Raveendran M; Hughes DST; Munzy DM; Yoder AD; Worley KC; Rogers J. 2021. Comparative genomic analysis of sifakas (Propithecus) reveals selection for folivory and high heterozygosity despite endangered status. Science Advances 7: eabd2274.
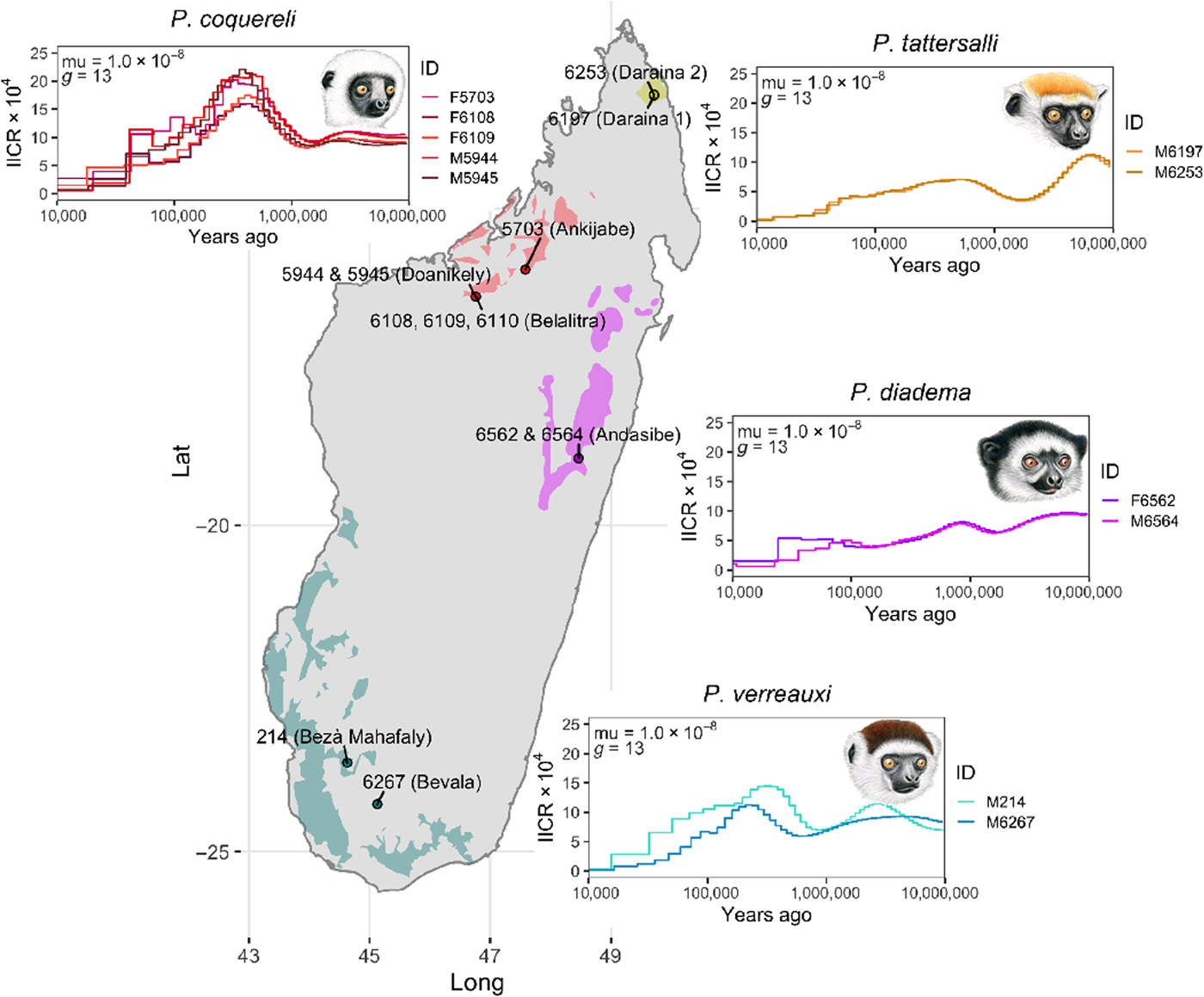
-
Access the paper
- Article Link
- Online Datasets:
19) Ozga AT*; Webster TH*; Gilby IC; Wilson MA; Nockerts RS; Wilson ML; Pusey AE; Li Y; Hahn BH; Stone AC. 2021. Urine as a high‐quality source of host genomic DNA from wild populations. Molecular Ecology Resources 21: 170-182.
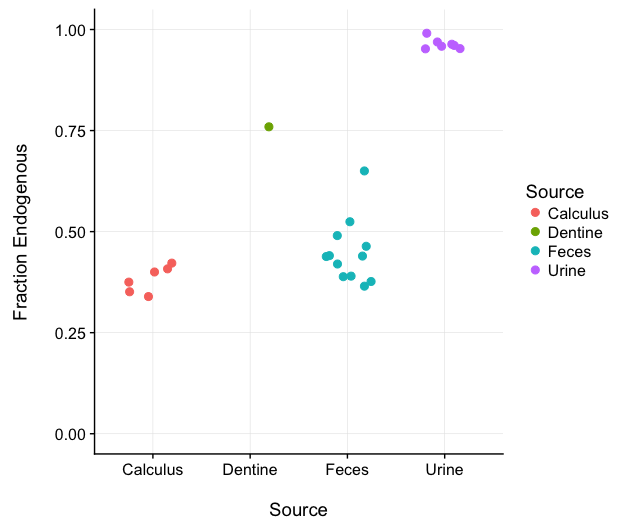
-
Access the paper
- Article Link
- Biorxiv Preprint: 955377
2020
18) Torosin NS; Argibay H; Webster TH; Corneli PS; Knapp LA. 2020. Comparing the selective landscape of TLR7 and TLR8 across primates reveals unique sites under positive selection in Alouatta. Molecular Phylogenetics and Evolution 152: 106920.
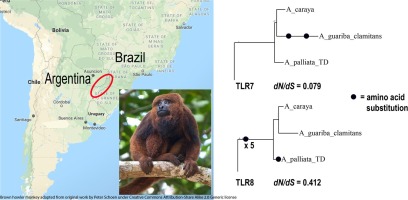
-
Access the paper
- Article Link
- Biorxiv Preprint: 708008 Additional Link:
- Press release: Silencing the booming chorus
17) Torosin NS; Webster TH; Argibay H; Ferreyra H; Uhart M; Agostini I; Knapp LA. 2020. Positively selected variants in functionally important regions of TLR7 in Alouatta guariba clamitans with yellow fever virus exposure in Northern Argentina. American Journal of Physical Anthropology 173: 50-60.
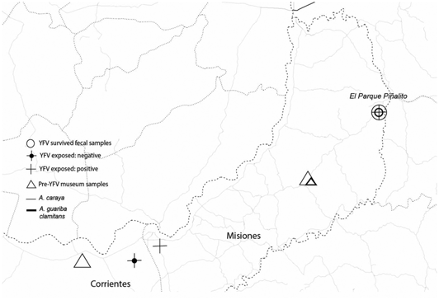
-
Access the paper
- Article Link
- Biorxiv Preprint: 725333 Additional Links:
- Press release: Silencing the booming chorus
- Press release: Mutaciones del ADN de los monos carayá rojos podrían haberlos salvado de la fiebre amarilla
16) Baden AL*; Webster TH*; Bradley BJ. 2020. Genetic relatedness cannot explain social preferences in black-and-white ruffed lemurs (Varecia variegata). Animal Behaviour 164: 73-82.
-
Access the paper
- Article Link
- Biorxiv Preprint: 799825
- Code: https://github.com/thw17/Varecia_social_preferences
15) Dolby GA; Morales M; Webster TH; DeNardo DF; Wilson MA; Kusumi K. 2020. Discovery of a new TLR gene and gene expansion event through improved desert tortoise genome assembly with chromosome-scale scaffolds. Genome Biology and Evolution 12: 3917-3925.
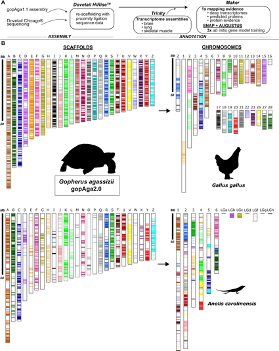
-
Access the paper
- Article Link
- Code: https://github.com/mmoral31/GopAga2.0-Genome
- Online Datasets:
14) Orton JP; Morales M; Fontenele RS; Schimidlin K; Kraberger S; Leavitt DJ; Webster TH; Wilson MA; Kusumi K; Dolby GA; Varsani A. 2020. Virus discovery in desert tortoise fecal samples: novel circular single stranded DNA viruses. Viruses 12: 143.
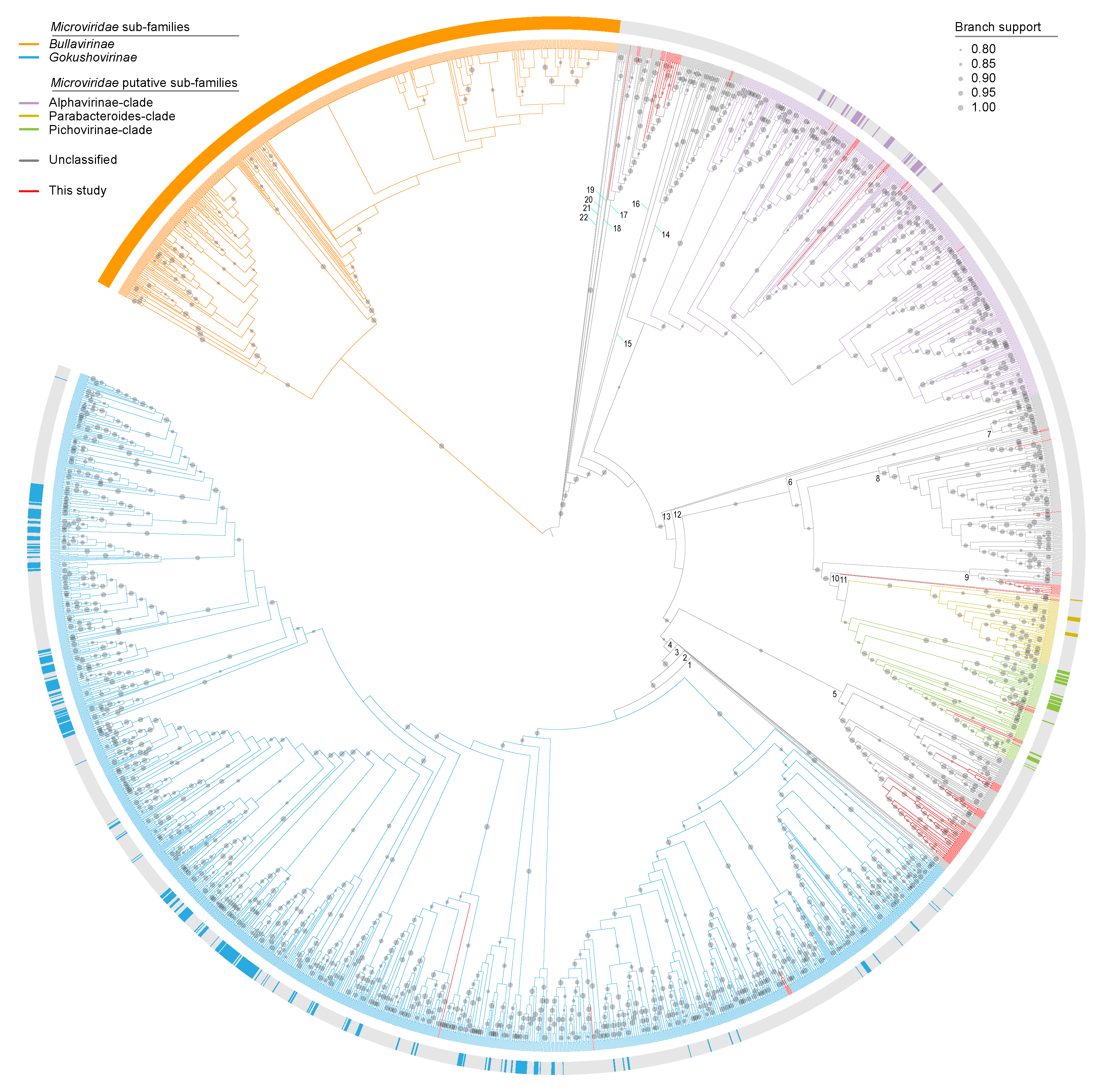
-
Access the paper
- Article Link
2019
13) Webster TH; Couse M; Grande BM; Karlins E; Phung TN; Richmond PA; Whitford W; Wilson Sayres MA. 2019. Identifying, understanding, and correcting technical artifacts on the sex chromosomes in next-generation sequencing data. Gigascience 8: giz074.

-
Access the paper
- Article Link
- Biorxiv Preprint: 346940
- Code: https://doi.org/10.5281/zenodo.2635885 Additional Link:
- XYalign Software Page
2018
12) Grüning B., et al. (164 total authors including Webster TH). 2018. Bioconda: a sustainable and comprehensive software distribution for the life sciences. Nature Methods 15: 475-476.

-
Access the paper
- Article Link
- Biorxiv Preprint: 207092
- Code: https://github.com/bioconda Additional Link:
- Bioconda Software Page
2017
11) Rupp S; Webster TH; Olney KC; Hutchins ED; Kusumi K; Wilson Sayres MA. 2017. Nearly complete dosage compensation in Anolis carolinensis, a reptile with XX/XY chromosomal sex determination. Genome Biology and Evolution 9: 231-240.
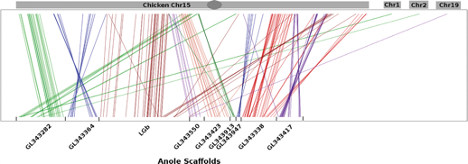
-
Access the paper
- Article Link
- Code: https://github.com/WilsonSayresLab/Anole_expression Additional Link:
- Code for diversity analyses
2016
10) Webster TH; Wilson Sayres MA. 2016. Genomic signals of sex-biased demography: progress and prospects. Current Opinions in Genetics and Development 41: 62-71.
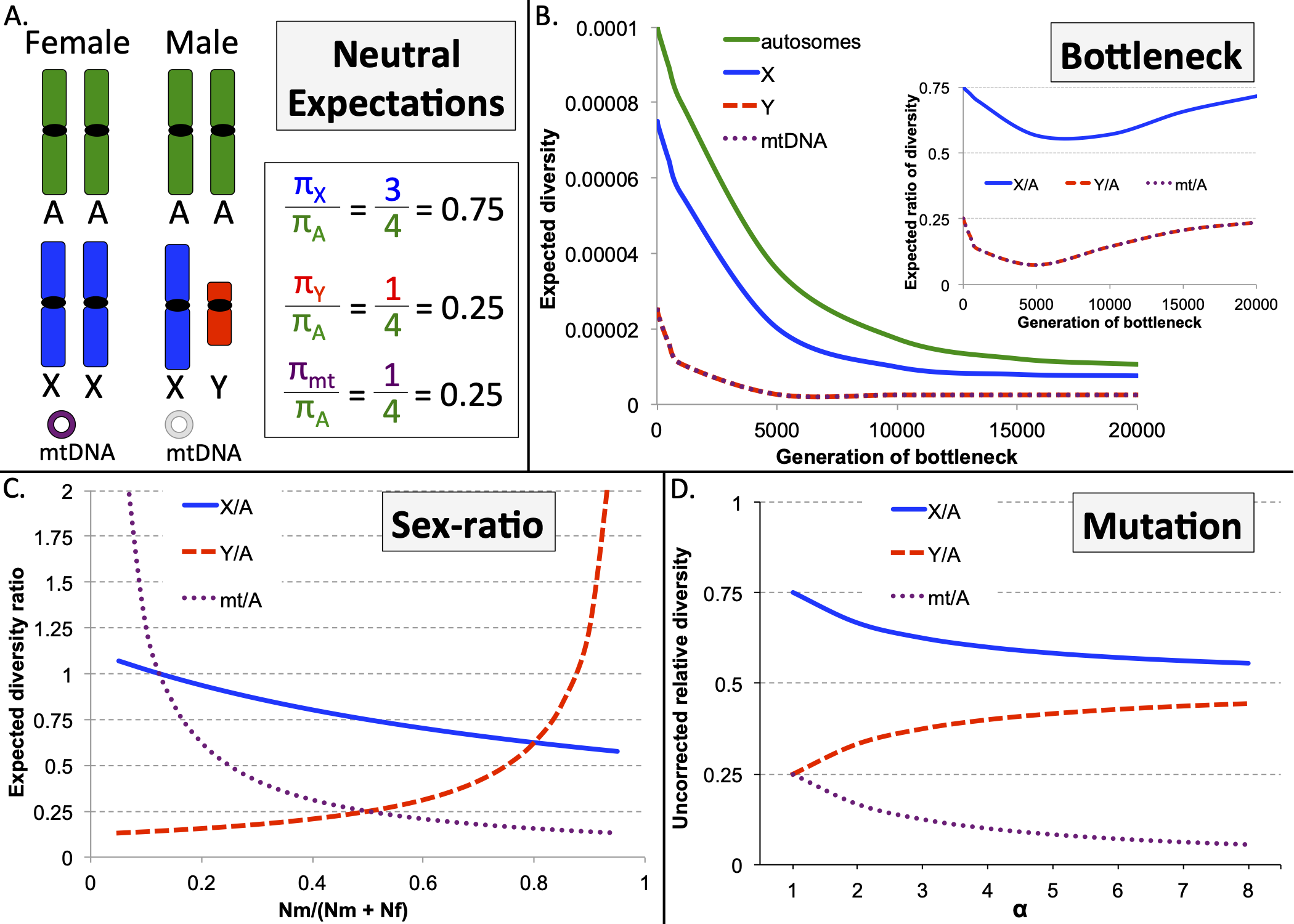
-
Access the paper
- Article Link
9) Baden AL; Webster TH; Kamilar JK. 2016. Resource seasonality and reproduction predict fission-fusion dynamics in a Malagasy strepsirrhine, Varecia variegata. American Journal of Primatology 78: 256-279.
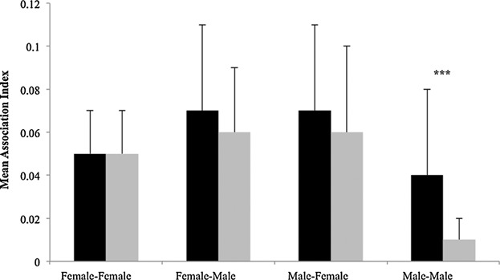
-
Access the paper
- Article Link
2015
8) Hsiang AY; Field DJ; Webster TH; Behlke ADB; Davis MB; Racicot RA; Gauthier JA. 2015. The origin of snakes: revealing the ecology, behavior, and evolutionary history of ancestral snakes using genomics, phenomics, and the fossil record. BMC Evolutionary Biology 15:87.

-
Access the paper
- Article Link
2014
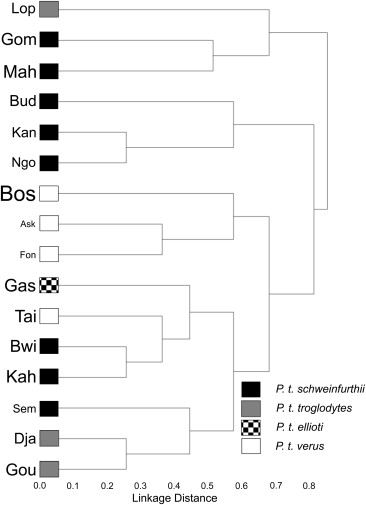
7) Webster TH; McGrew WC; Marchant LF; Payne CLR; Hunt KD. 2014. Selective insectivory at Toro-Semliki, Uganda: Comparative analyses suggest no ‘savanna’ chimpanzee pattern. Journal of Human Evolution 71: 20-27.
-
Access the paper
- Article Link
6) Anestis SF; Webster TH; Kamilar JK; Fontenot MB; Watts DP; Bradley BJ. 2014. AVPR1a variation in chimpanzees (Pan troglodytes): population differences and association with behavioral style. International Journal of Primatology 35(1): 305-324.

-
Access the paper
- Article Link
2013
5) McGrew WC; Marchant LF; Payne CLR; Webster TH; Hunt KD. 2013. Well digging by Semliki chimpanzees: new data on laterality and hydrology. Pan Africa News 20(1): 5-8.

-
Access the paper
- Article Link
2012
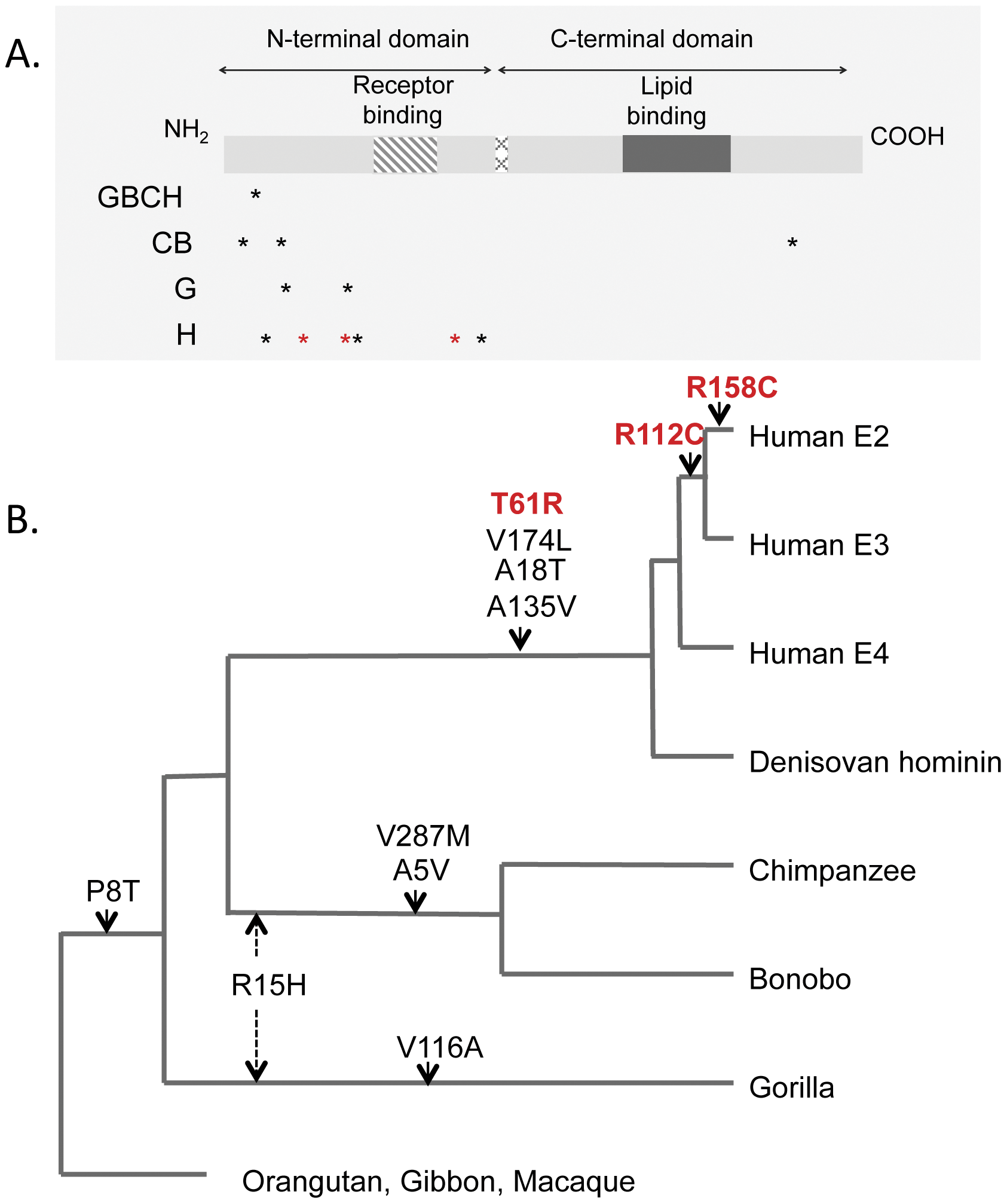
4) McIntosh AM; Bennett C; Dickson D; Anestis SF; Watts DP; Webster TH; Fontenot MB; Bradley BJ. 2012. The apolipoprotein E (APOE) gene appears functionally monomorphic in chimpanzees (Pan troglodytes). PLoS ONE 7(10): e47760.
-
Access the paper
- Article Link
2010
3) McGrew WC; Marchant LF; Payne CLR; Webster TH; Hunt KD. 2010. Chimpanzees at Semliki ignore oil palms. Pan Africa News 17(2): 19-21.

-
Access the paper
- Article Link
2009
2) Webster TH; Hodson PR; Hunt KD. 2009. Grooming hand-clasp by chimpanzees of the Mugiri community, Toro-Semliki Wildlife Reserve, Uganda. Pan Africa News 16 (1): 5-7.

-
Access the paper
- Article Link
2008
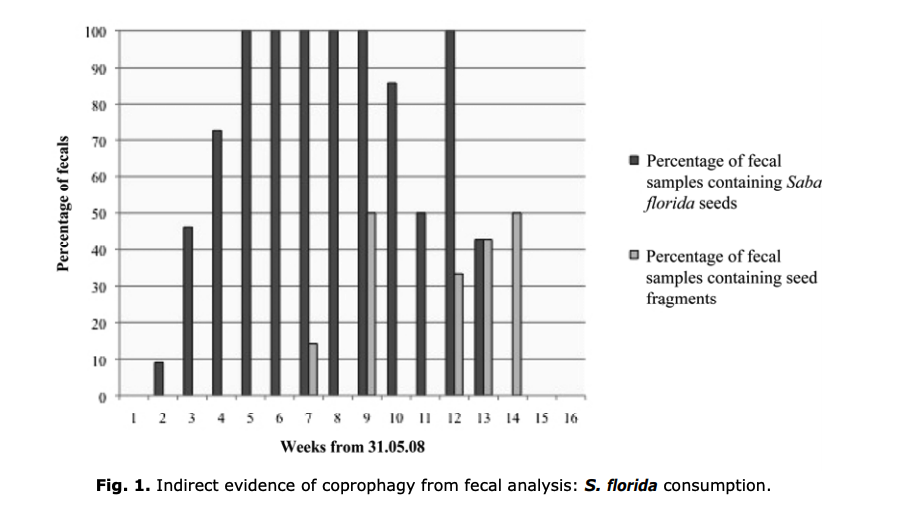
1) Payne CLR; Webster TH; Hunt KD. 2008. Coprophagy by the semi-habituated chimpanzees of Semliki, Uganda. Pan Africa News 15 (2): 29-32.
-
Access the paper
- Article Link